Dr. Ivet Bahar
Louis and Beatrice Laufer Endowed Chair
Director, Laufer Center for Physical and Quantitative Biology
Professor, Department of Biochemistry and Cell Biology
Stony Brook University, Renaissance School of Medicine
Phone: (631) 632-5422
E-mail: ivet.bahar@stonybrook.edu

- Research Description
Biomolecular Systems Dynamics
The dynamics of living systems at the molecular level defines cell survival and regulation mechanisms, which in turn, define our physiological responses. Biomolecules work together in the cell in a seamless way: they communicate (intra- and intermolecular signaling), interact with each other (chemical reactions, binding, complex formation, assembly), produce and consume energy (mitochondrial functions, metabolic events), carry cargos (ion or substrate transport). They not only sustain and regulate cell life (transcription, translation, repair), but also orderly terminate it (apoptosis). How do biomolecules achieve so many functions? Because they have flexible structures that can adapt to changes and act in specific ways (intrinsic dynamics), optimized by evolution to sustain their activities, driven by internal or external stimuli, controlled by principles of physical sciences and engineering. Chemical and physical rules that apply at the microscopic scale are not any different from those of the macroscopic world. Our goal is to help improve our understanding of the basic principles of these actions with the help of computing technology, visualize/simulate their time evolution, and discover rational intervention methods to counter their dysfunction.
Schematic representation of relationships between mechanisms of action, evolution and dynamics. Green round box represents the mechanisms of action. If an action is functional, it has allosteric and/or orthosteric effects. If it is dysfunctional, it will not be evolutionarily selected (Blue round box). The protein sequences and structures filtered by evolution perform their functions defined by intrinsic dynamics, during which perturbations act through either physical or chemical changes (orange round box). These changes influence the selection of modes of motion, and thereby actions. Gray square boxes represent four main stimuli that trigger allosteric responses. (Ref: Zhang Y, Doruker P, Kaynak B, Zhang S, Krieger JM, Li H, Bahar I. (2020) Intrinsic dynamics is evolutionarily optimized to enable allosteric behavior. Curr Opin Struct Biol 62:14-21.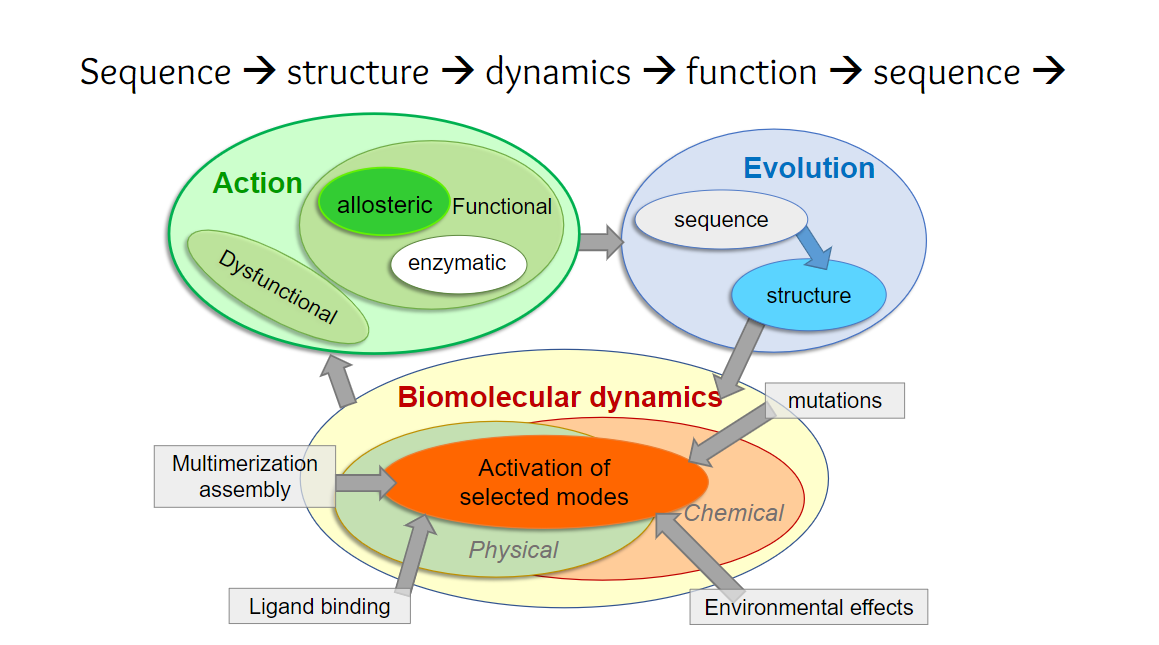
- Selected Publications
PUBLICATIONS PAGE | NCBI LIST | CITATIONS
Publications 2024
Carlos Ventura, Anupam Banerjee, Maria Zacharopoulou, Laura S Itzhaki, Ivet Bahar (2024) Tandem-repeat proteins conformational mechanics are optimized to facilitate functional interactions and complexations. Current Opinion in Structural Biology 84, 102744
J371. MGS Costa, M Gur, JM Krieger, Bahar I (2023) Computational biophysics meets cryo-EM revolution in the search for the functional dynamics of biomolecular systems. Wiley Interdisciplinary Reviews: Computational Molecular Science, e1689
J370. Valerian E Kagan, Yulia Y Tyurina, Karolina Mikulska-Ruminska, Deena Damschroder, Eduardo Vieira Neto, Alessia Lasorsa, Alexander A Kapralov, Vladimir A Tyurin, Andrew A Amoscato, Svetlana N Samovich, Austin B Souryavong, Haider H Dar, Abu Ramim, Zhuqing Liang, Pablo Lazcano, Jiajia Ji, Michael W Schmidtke, Kirill Kiselyov, Aybike Korkmaz, Georgy K Vladimirov, Margarita A Artyukhova, Pushpa Rampratap, Laura K Cole, Ammanamanchi Niyatie, Emma-Kate Baker, Jim Peterson, Grant M Hatch, Jeffrey Atkinson, Jerry Vockley, Bernhard Kühn, Robert Wessells, Patrick CA van der Wel, Bahar I, Hülya Bayir, Miriam L Greenberg (2023) Anomalous peroxidase activity of cytochrome c is the primary pathogenic target in Barth syndrome. Nature Metabolism 5, 2184-2205
J369. Denys Bondar, Olga Bragina, Ji Young Lee, Ivan Semenyuta, Ivar Järving, Volodymyr Brovarets, Peter Wipf, Bahar I, Yevgen Karpichev (2023) Hydroxamic Acids as PARP-1 Inhibitors: Molecular Design and Anticancer Activity of Novel Phenanthridinones. Helvetica Chimica Acta 106 (10), e202300133
J368. Yan Zhang, Xiaojie Qiu, Jonathan S Weissman, Ivet Bahar, Jianhua Xing (2023) Graph-Dynamo: Learning stochastic cellular state transition dynamics from single cell data. BioRxiv, 2023.09.24.559170
J367. MP Dalton, MH Cheng, Bahar I, JA Coleman (2023) Structural Mechanisms for VMAT2 inhibition by tetrabenazine . BioRxiv 2023.09.05.556211
P2. Bahar I, H Cheng, S Zhang, R Porritt, M Arditi (2023) Compositions and methods for treating a COVID-19 infection . US Patent 11,746,144
J366. Dar HH, Mikulska-Ruminska K, Tyurina YY, Luci DK, Yasgar A, Samovich SN, Kapralov AA, Souryavong AB, Tyurin VA, Amoscato AA, Epperly MW, Shurin GV, Standley M, Holman TR, St. Croix CM, Watkins SC, VanDemark AP, Rana S, Zakharov AV, Simeonov A, Marugan J, Mallampalli RK, Wenzel SE, Greenberger JS, Rai G, Bayir H, Bahar I, Kagan VE (2023) Discovering selective antiferroptotic inhibitors of the 15LOX/PEBP1 complex noninterfering with biosynthesis of lipid mediators. Proc Natl Acad Sci (USA) 120, e2218896120
J365. Zhang C, Zhang X, Weiß T, Cheng MH, Chen S, Ambrosius CK, Czerniak AS, Li K, Feng M, Bahar I, Beck-Sickinger AG (2023) Structural basis of CMKLR1 signaling induced by chemerin9. BioRxiv , 2023.06. 09.544295
J364. Amoscato AA, Anthonymuthu T, Kapralov O, Sparvero LJ, Shrivastava IH, Mikulska-Ruminska K, Tyurin VA, Shvedova AA, Tyurina YY, Bahar I, Wenzel S, Bayir H, Kagan VE (2023) Formation of protein adducts with hydroperoxy-PE electrophilic cleavage products during ferroptosis. Redox Biology 63, 1-17, 102758
J363. Banerjee A, Bahar I (2023) Structural Dynamics Predominantly Determine the Adaptability of Proteins to Amino Acid Deletions. International Journal of Molecular Sciences 24 (9), 8450
J362. Kaynak BT, Dahmani ZL, Doruker P, Banerjee A, Yang SH, Gordon R, Itzhaki LS, Bahar I (2023) Cooperative mechanics of PR65 scaffold underlies the allosteric regulation of the phosphatase PP2A. Structure 31, 607-618.
J361. Banerjee A, Saha S, Tvedt NC, Yang LW, Bahar I (2023) Mutually beneficial confluence of structure-based modeling of protein dynamics and machine learning methods. Current Opinion in Structural Biology 78, 102517
J360. Shekar A, Mabry SJ, Cheng MH, Aguilar JI, Patel S, Zanella D, Saleeby DP, Zhu Y, Romanazzi T, Ulery-Reynolds P, Bahar I, Angela M Carter, Heinrich JG Matthies, Aurelio Galli (2023) Syntaxin 1 Ser14 phosphorylation is required for nonvesicular dopamine release Science Advances, 9(2), eadd8417
J359. Vilardaga JP, Clark LJ, White AD, Sutkeviciute I, Lee JY, Bahar I (2023) Molecular Mechanisms of PTH/PTHrP class B GPCR Signaling and Pharmacological Implications. Endocrine Reviews, 44, 474-491. https://doi.org/10.1210/endrev/bnac032
J358. Noval Rivas M, Porritt RA, Cheng MH, Bahar I M Arditi (2022) Multisystem inflammatory syndrome in children and long COVID: the SARS-CoV-2 viral superantigen hypothesis. Frontiers in Immunology, 3480
J357. Smith IN, Dawson JE, Krieger J, Thacker S, Bahar I, Eng C (2022) Structural and Dynamic Effects of PTEN C-Terminal Tail Phosphorylation. J Chem Inf Model. 62(17):4175-4190. PMID: 36001481
J356. Abram A, Jakubiec M, Reeb K, Cheng MH, Gedschold R, Rapacz A, Mogilski S, Socala K, Nieoczym D, Szafarz M, Latacz G, Szulczyk B, Kalinowska-Tluscik J, Gawel K, Esguerra CV, Wyska E, Müller CE, Bahar I, Fontana ACK, Wlaz P, Kaminski RM, Kaminski K (2022) Discovery of (R)-N-Benzyl-2-(2,5-dioxopyrrolidin-1-yl)propanamide [(R)-AS-1], a Novel Orally Bioavailable EAAT2 Modulator with Drug-like Properties and Potent Antiseizure Activity In Vivo. J. Med. Chem. 65, 17, 11703-11725
J355. Todd NK, Huang Y, Lee JY, Doruker P, Krieger JM, Salisbury R, MacDonald M, Bahar I, Thathiah A (2022) GPCR kinases generate an APH1A phosphorylation barcode to regulate amyloid-β generation Cell Reports 40 (3), 111110
J354. Wingert B, Doruker P, Bahar I (2022). Activation and Speciation Mechanisms in Class A GPCRs. J Mol Biol. 434 (17), 167690.
J353. Lefever DE, Miedel MT, Pei F, DiStefano JK, Debiasio R, Shun TY, Saydmohammed M, Chikina M, Vernetti LA, Soto-Gutierrez A, Monga SP, Bataller RA, Behari J, Yechoor VK, Bahar I, Gough A, Stern AM, Taylor DL (2022) A Quantitative Systems Pharmacology Platform Reveals NAFLD Pathophysiological States and Targeting Strategies. Metabolites 12(6), 528
J352. Acar O, Zhang S, Bahar I, Carvunis AR (2022) Elastic Network Modeling of Cellular Networks Unveils Sensor and Effector Genes that Control Information Flow. PLOS Computational Biology 18 (5), e1010181. PMID: 35639793
J351. Krieger JM, Sorzano COS, Carazo JM, Bahar I (2022) Protein dynamics developments for the large scale and cryoEM: case study of ProDy 2.0. Acta Crystallographica Section D: Structural Biology 78 (4) PMID: 35362464 PMCID: PMC8972803
J350. Colas C, Bahar I, Shi L, Font Sadurni J (2022) Editorial: Targeting Membrane Proteins: Structure-Function-Dynamics Relationships. Frontiers in Chemistry, 126PMID: 35308796
J349. Refai O, Aggarwal S, Cheng MH, Gichi Z, Salvino JM, Bahar I, Blakely RD, Mortensen OV (2022) Allosteric Modulator KM822 Attenuates Behavioral Actions of Amphetamine in Caenorhabditis elegans through Interactions with the Dopamine Transporter DAT-1. Molecular Pharmacology 101 (3) 123-131. PMID: 34906999 PMCID: PMC8969146
J348. Sang Z, Xiang Y, Bahar I, Shi Y. (2022) Llamanade : an open-source computational pipeline for robust nanobody humanization.. Structure, 30(3):418-429.e3. PMID: 34373858 PMCID: PMC8351782
J347. Sutkeviciuta I, Lee JY, White AD, Maria CS, Peña KA, Savransky S, Doruker P, Li H, Lei S, Kaynak B, Tu C, Clark LJ, Sanker S, Gardella TJ, Chang W, Bahar I* and Vilardaga JP*. (2022) Precise druggability of the PTH type 1 receptor. Nature Chem Biol. 18(3):272-280. PMID: 34949836 PMCID: PMC8891041
J346. Cheng MH, Krieger JM, Banerjee A, Xiang Y, Kaynak B, Shi Y, Arditi M, Bahar I (2022) Impact of new variants on SARS-CoV-2 infectivity and neutralization: A molecular assessment of the alterations in the spike-host protein interactions. iScience 25(3):103939. PMID: 35194576 PMCID: PMC8851820
J345. Chen C, Saville JW, Marti MM, Schäfer A, Cheng MH, Mannar D, Zhu X, Berezuk AM, Banerjee A, Sobolewski MD, Kim A, Treat BR, Da Silva Castanha PM, Enick N, McCormick KD, Liu X, Adams C, Hines MG, Sun Z, Chen W, Jacobs JL, Barratt-Boyes SM, Mellors JW, Baric RS, Bahar I, Dimitrov DS, Subramaniam S, Martinez DR, Li W (2022) Potent and Broad Neutralization of SARS-CoV-2 Variants of Concern (VOCs) including Omicron Sub-lineages BA. 1 and BA. 2 by Biparatopic Human VH Domains. iScience 25(8), 104798
J344. Qiu X, Zhang Y, Martin-Rufino JD, Weng C, Hosseinzadeh S, Yang D, Pogson AN, Hein MY, Hoi Joseph Min K, Wang L, Grody EI, Shurtleff MJ, Yuan R, Xu S, Ma Y, Replogle JM, Lander ES, Darmanis S, Bahar I, Sankaran VG, Xing J, Weissman JS (2022) Mapping transcriptomic vector fields of single cells. Cell 185:690-711. PMID: 35108499
J343. Lamade AM, Wu L, Dar H, Mentrup HL, Shrivastava IH, Epperly MW, St Croix CM, Tyurina YY, Anthonymuthu TS, Yang Q, Kapralov AA, Huang Z, Mao G, Amoscato AA, Hier ZE, Artyukova MA, Shurin G, Rosenbaum JC, Gough PJ, Bertin J, VanDemark AP, Watkins SC, Mollen KP, Bahar I, Greenberger JS, Kagan VE, Whalen MJ, Bayιr H. (2022) Inactivation of RIP3 kinase sensitizes to 15LOX/PEBP1-mediated ferroptotic death. Redox Biology 50, 102232 PMID: 35101798 PMCID: PMC8804265
J342. Kaynak BT, Krieger JM, Dudas B, Dahmani ZL, Costa MGS, Balog E, Scott AL, Doruker P, Perahia D, Bahar I. (2022) Sampling of Protein Conformational Space Using Hybrid Simulations: A Critical Assessment of Recent Methods. Front Mol Biosci., 9:832847 PMID: 35187088 PMCID: PMC8855042
J341. Calero DH, Lederman R, Krieger J, Myška D, Strelak D, Filipovic J, Bahar I, Carazo JM, Sorzano CO. (2021) Continuous heterogeneity analysis of CryoEM images through Zernike polynomials and spherical harmonics. Microscopy and Microanalysis, 27 (S1), 1680-1682
J340. Romanazzi T, Zanella D, Cheng MH, Smith B, Carter AM, Galli A, Bahar I, Bossi E. (2021) Bile acids gate dopamine transporter mediated currents. Frontiers in Chemistry 9 PMID: 34957043 PMCID: PMC8702627
J339. Herreros D, Lederman RR, Krieger J, Jiménez-Moreno A, Martínez M, Myška D, Strelak D, Filipovic J, Bahar I, Carazo JM, Sanchez COS. (2021) Approximating deformation fields for the analysis of continuous heterogeneity of biological macromolecules by 3D Zernike polynomials. IUCrJ 8(Pt 6):992-1005. PMID: 34804551 PMCID: PMC8562670
J338. White AD, Pena KA, Clark LJ, Maria CS, Liu S, Jean-Alphonse FG, Lee JY, Lei S, Cheng Z, Tu CL, Fang F, Szeto N, Gardella TJ, Xiao K, Gellman SH, Bahar I, Sutkeviciute I, Chang W, Vilardaga JP. (2021) Spatial Bias in cAMP Generation Determines Biological Responses to PTH type 1 Receptor Activation. Sci Signal. 14(703):eabc5944. PMID: 34609896 PMCID: PMC8682804
J337. Chen F, Shi Q, Pei F, Vogt A, Porritt RA, Garcia G, Gomez AC, Cheng MH, Schurdak ME, Liu B, Chan SY, Arumugaswami V, Stern AM, Taylor DL, Arditi M, Bahar I. (2021) A Systems-level Study Reveals Host-targeted Repurposable Drugs Against SARS-CoV2 Infection. Molecular Systems Biol. 17:e10239 PMID: 34339582 PMCID: PMC8328275
J336. Kaynak BT, Zhang S, Bahar I, Doruker P (2021) ClustENMD: Efficient sampling of biomolecular conformational space at atomic resolution. Bioinformatics 37:3956-8. PMID: 34240100 PMCID: PMC8570821
J335. Zhang Y, Zhang S, Xing J, Bahar I (2021) Normal mode analysis of membrane protein dynamics using the vibrational subsystem analysis. J Chem Phys 154, 195102 PMID: 34240914 PMCID: PMC8131107
J334. Mikulska-Ruminska K, Anthonymuthu TS, Levkina A, Shrivastava IH, Kapralov AA, Bayir H, Kagan VE, Bahar I. (2021) NO● Represses the Oxygenation of Arachidonoyl PE by 15LOX/PEBP1: Mechanism and Role in Ferroptosis. Int J Mol Sci. 22(10): 5253. PMID: 34067535 PMCID: PMC8156958
J333. Aggarwal J, Cheng MH, Salvino JM, Bahar I, Mortensen OV. (2021) Functional Characterization of the Dopaminergic Psychostimulant Sydnocarb as an Allosteric Modulator of the Human Dopamine Transporter. Biomedicines, 9(6), 634; https://doi.org/10.3390/biomedicines9060634 PMID: 34199621 PMCID: PMC8227285
J332. Aguilar JI, Cheng MH, Font J, Schwartz AC, Ledwitch K, Duran A, Mabry SJ, Belovich AN, Zhu Y, Carter AM, Shi L, Kurian MA, Fenollar-Ferrer C, Meiler J, Ryan RM, Mchaourab HS, Bahar I, Matthies HJG, Galli A. (2021) Psychomotor impairments and therapeutic implications revealed by a mutation associated with infantile Parkinsonism-Dystonia. ELife, 10, e68039 PMID: 34002696 PMCID: PMC8131106
J331. Cheng MH, Porritt RA, Rivas MN, Krieger JM, Ozdemir AB, Garcia G Jr, Arumugaswami V, Fries BC, Arditi M, Bahar I. (2021) A monoclonal antibody against staphylococcal enterotoxin B superantigen inhibits SARS-CoV-2 entry in vitro. Structure. 29(9):951-962 PMID: 33269352 PMCID: PMC7709177
J330. Zhang S, Krieger JM, Zhang Y, Kaya C, Kaynak B, Mikulska-Ruminska K, Doruker P, Li H, Bahar I. (2021) ProDy 2.0: Increased Scale and Scope after 10 Years of Protein Dynamics Modelling with Python. Bioinformatics 37:3657-9 PMID: 33822884 PMCID: PMC8545336
J329. Pei F, Shi Q, Zhang H, Bahar I (2021) Predicting Protein-Protein Interactions Using Symmetric Logistic Matrix Factorization. J Chem Inf Model 61:1670-1682. PMID: 33831302 PMCID: PMC8253547
J328. Porritt RA, Paschold L, Noval Rivas M, Cheng MH, Yonker LM, Chandnani H, Lopez M, Simnica D, Schultheiß C, Santiskulvong C, van Eyk J, McCormick JK, Fasano A, Bahar I, Binder M, Arditi M (2021) HLA class I-associated expansion of TRBV11-2 T cells in Multisystem Inflammatory Syndrome in Children. J Clin Invest. 131(10). PMID: 33705359 PMCID: PMC8121516 Link to article /commentary
J327. Sorkina T, Cheng MH, Bagalkot TR, Wallace C, Watkins SC, Bahar I, Sorkin A. (2021) Direct coupling of oligomerization and oligomerization-driven endocytosis of the dopamine transporter to its conformational mechanics and activity. J Biol Chem. Feb 18:100430. PMID: 33610553 PMCID: PMC8010718
J326. Wang H, Pei F, Vanyukov MM, Bahar I, Wu W, Xing EP. (2021) Coupled mixed model for joint genetic analysis of complex disorders with two independently collected data sets. BMC Bioinformatics 22(1):50. PMID: 33546598 PMCID: PMC7866684
J325. Sun WY, Tyurin VA, Mikulska-Ruminska K, Shrivastava IH, Anthonymuthu TS, Zhai YJ, Pan MH, Gong HB, Lu DH, Sun J, Duan WJ, Korolev S, Abramov AY, Angelova PR, Miller I, Beharier O, Mao GW, Dar HH, Kapralov AA, Amoscato AA, Hastings TG, Greenamyre TJ, Chu CT, Sadovsky Y, Bahar I, Bayir H, Tyurina YY, He RR, Kagan VE. (2021) Phospholipase iPLA2ß averts ferroptosis by eliminating a redox lipid death signal. Nat Chem Biol 17(4):465-476. PMID: 33542532 PMCID: PMC8152680
J324. Meirovitch H, Bahar I. (2021) Harold A. Scheraga (10/18/1921—8/1/2020): A pioneering scientist who laid the foundations of protein science in the 20th century. Proc Natl Acad Sci USA 118(8):e2026796118. PMID: 33547253 PMCID: PMC7923550.
J323. Cheng MH, Krieger JM, Kaynak B, Arditi M, Bahar I. (2021) Impact of South African 501.V2 Variant on SARS-CoV-2 Spike Infectivity and Neutralization: A Structure-based Computational Assessment. BioRxiv [Online ahead of print].
J322. Zheng F, Zhang S, Churas C, Pratt D, Bahar I, Ideker T (2021) HiDeF: Identifying Persistent Structures in Multiscale 'Omics Data. Genome Biol. 22: 21. PMID: 33413539 PMCID: PMC7789082
J321. Dagvadorj J, Mikulska-Ruminska K, Tumurkhuu G, Ratsimandresy RA, Carriere J, Andres AM, Marek-Iannucci S, Song Y, Chen S, Lane M, Dorfleutner A, Gottlieb RA, Stehlik C, Cassel S, Sutterwala FS, Bahar I, Crother TR, Arditi M (2021) Recruitment of pro-IL-1a to Mitochondrial Cardiolipin, via Shared LC3 Binding Domain, Inhibits Mitophagy and Drives Maximal NLRP3 Activation. Proc Natl Acad Sci USA 118:e2015632118 PMID: 33361152 PMCID: PMC7817159
J320. Rivas MN, Porritt RA, Cheng MH, Bahar I, Arditi M. (2021) COVID-19 Associated Multisystem Inflammatory Syndrome in Children (MIS-C): A Novel Disease that Mimics Toxic Shock Syndrome. The Superantigen Hypothesis. J Allergy Clin Immunol. 147: 57-59. PMID: 33075409 PMCID: PMC7564502
J319. Anthonymuthu TS, Tyurina YY, Sun WY, Mikulska-Ruminska K, Shrivastava IH, Tyurin VA, Cinemre FB, Dar HH, VanDemark AP, Holman TR, Sadovsky YS, Stockwell BR, He RR, Bahar I, Bayir H, Kagan VE. (2021) Resolving the Paradox of Ferroptotic Cell Death: Ferrostatin-1 Binds to 15LOX/PEBP1 Complex, Suppresses Generation of Peroxidized ETE-PE, and Protects against Ferroptosis. Redox Biol. 38: 101744 PMID: 33126055 PMCID: PMC7596334 .
J318. Wingert B, Krieger JM, Li H, Bahar I (2021) Adaptability and Specificity: How Do Proteins Balance Opposing Needs to Achieve Function? Curr Opin Struct Biol 67:25-32. PMID: 33053463 PMCID: PMC8036234
J317. Zhang Y, Krieger JM, Mikulska-Ruminska K, Kaynak B, Sorzano COS, Carazo JM, Xing J, Bahar I (2021) State-dependent sequential allostery exhibited by chaperonin TRiC/CCT revealed by network analysis of Cryo-EM maps. Prog Biophys Mol Biol S0079-6107(20)30082-1. PMID: 32866476 PMCID: PMC7914283
J316. Belovich AN, Aguilar JI, Mabry SJ, Cheng MH, Zanella D, Hamilton PJ, Stanislowski DJ, Shekar A, Foster JD, Bahar I, Matthies HJG, Galli A. (2021) A network of phosphatidylinositol (4, 5)-bisphosphate (PIP 2) binding sites on the dopamine transporter regulates amphetamine behavior in Drosophila Melanogaster. Molecular Psychiatry 26, 4417-4430. PMID: 31796894 PMCID: PMC7266731
J315. Porritt RA, Paschold L, Rivas MN, Cheng MH, Yonker LM, Chandnani H, Lopez M, Simnica D, Schultheiβ C, Santiskulvong C, Van Eyk J, Fasano A, Bahar I, Binder M, Arditi M. (2020) Identification of a Unique TCR Repertoire, Consistent with a Superantigen Selection Process in Children with Multi-system Inflammatory Syndrome. BioRxiv doi: https://doi.org/10.1101/2020.11.09.372169
J314. Cheng MH, Porritt RA, Rivas MN, Krieger JM, Ozdemir AB, Garcia G, Arumugaswami V, Fries BC, Arditi M, Bahar I. (2020) A Monoclonal Antibody Against Staphylococcal Enterotoxin B Superantigen Inhibits SARS-CoV-2 entry in vitro. BioRxiv doi: https://doi.org/10.1101/2020.11.24.395079
J313. Zhou Z, Huang F, Shrivastava IH, Zhu R, Luo A, Hottiger M, Bahar I, Liu Z, Cristofanilli M, Wan Y (2020) New Insight into the Significance of KLF4 PARylation in Genome Stability, Carcinogenesis, and Therapy. EMBO Molecular Medicine, 12(12): e12391.
J312. Song X, Zhou Z, Li H, Xue Y, Lu X, Bahar I, Kepp O, Hung MC, Kroemer G, Wan Y (2020) Pharmacological Suppression of B7-H4 Glycosylation Restores Antitumor Immunity in Immune-Cold Breast Cancers. Cancer Discov 10:1872-1893. doi: 10.1158/2159-8290.CD-20-0402. [Online ahead of print]. PMID: 32938586 PMCID: 7710601
J311. Cheng MH, Zhang S, Porritt RA, Rivas MN, Paschold L, Willscher E, Binder M, Arditi M*, Bahar I* (2020) Superantigenic character of an insert unique to SARS-CoV-2 spike supported by skewed TCR repertoire in patients with hyperinflammation. Proc Natl Acad Sci USA 117: 25254-25262 PMID: 32989130 PMCID: PMC7568239 US Patent application: Compositions and methods for treating a covid-19 infection I Bahar, H Cheng, S Zhang, R Porritt, M Arditi US Patent App. 17/375,601
J310. Clark LJ, Krieger J, White AD, Bondarenko V, Lei S, Fang F, Lee JY, Doruker P, Böttke T, Jean-Alphonse F, Tang P, Gardella TJ, Xiao K, Sutkeviciute I, Coin I, Bahar I, Vilardaga JP (2020) Allosteric interactions in the parathyroid hormone GPCR-arrestin complex formation. Nat Chem Biol 16(10):1096-1104 PMID: 32632293 PMCID: PMC7502484 (available on 2021-01-06)
J309. Kaynak BT,* Bahar I,* Doruker P. (2020) Essential Site Scanning Analysis: a new approach for detecting sites that modulate the dispersion of protein global motions. Comp Struct Biotech J 18:1577-1586. PMID: 32637054 PMCID: PMC7330491
J308. Krieger JM, Doruker P, Scott AL, Perahia D, Bahar I. (2020) Towards Gaining Sight of Multiscale Events: Utilizing Network Models and Normal Modes in Hybrid Methods. Curr Opin Struct Biol 64:34-41. PMID: 32622329 PMCID: PMC7666066 (available on 2021-10-01)
J307. Zheng F, Zhang S, Churas C, Pratt D, Bahar I, Ideker T. (2020) Decoding of persistent multiscale structures in complex biological networks. bioRxiv. PMID: 32587977 PMCID: PMC7310637
J306. Zhao J, Dar HH, Deng Y, St. Croix CM, Li Z, Minami Y, Shrivastava IH, Tyurina YY, Etling E, Rosenbaum JC, Nagasaki T, Trudeau JB, Watkins SC, Bahar I, Bayir H, VanDemark AP, Kagan VE, Wenzel SE. (2020) PEBP1 Acts as a Rheostat Between Prosurvival Autophagy and Ferroptotic Death in Asthmatic Epithelial Cells. Proc Natl Acad Sci (USA) 117:14376-14385. PMID: 32513718 PMCID: PMC7321965
J305. Kim Y, Jun I, Shin DH, Yoon JG, Piao H, Jung J, Park HW, Cheng MH, Bahar I, Whitcomb DC, Lee MG (2020) Regulation of CFTR Bicarbonate Channel Activity by WNK1: Implications for Pancreatitis and CFTR-Related Disorders. Cell Mol Gastroenterol Hepatol 9:71-103. PMID: 31561038 PMCID: PMC6889609
J304. Shi Q, Pei F, Silverman GA, Pak SC, Perlmutter DH, Liu B*, Bahar I*. (2020) Mechanisms of Action of Autophagy Modulators Dissected by Quantitative Systems Pharmacology Analysis. Int. J. Mol. Sci 21:2855. PMID: 32325894 PMCID: PMC7215584
J303. Ponzoni L, Nguyen NH, Bahar I*, Brodsky JL*. (2020) Complementary Computational and Experimental Evaluation of Missense Variants in the ROMK Potassium Channel. PLOS Comp Biol 16, e1007749. PMID: 32251469 PMCID: PMC7162551
J302. Li H, Doruker P, Hu G, Bahar I. (2020) Modulation of Toroidal Proteins Dynamics in Favor of Functional Mechanisms upon Ligand Binding. Biophys J 118: 1782-1794. PMID: 32130874 PMCID: 7136436
J301. Li H, Pei F, Taylor DL, Bahar I. (2020) QuartataWeb: Integrated Chemical-protein-pathway Mapping for Polypharmacology and Chemogenomics. Bioinformatics 36:3935-3937. PMID: 32221612 PMCID: PMC7320630
J300. Zhou Z, Zhou H, Ponzoni L, Luo A, Zhu R, He M, Huang Y, Guan KL, Bahar I, Liu Z, Wan Y. (2020) EIF3H Orchestrates Hippo Pathway-mediated Oncogenesis Through an Unexpected Catalytic Control of YAP Stability. Cancer Res 80:2550-2563. PMID: 32269044 PMCID: PMC7316131
J299. Cheng MH, Zhang S, Porritt RA, Arditi M, Bahar I. (2020) An insertion unique to SARS-CoV-2 exhibits superantigenic character strengthened by recent mutations. BioRxiv. 2020 May 21:2020.05.21.109272. PMID: 32511374 PMCID: PMC7263503
J298. Thermozier S, Hou W, Zhang X, Shields D, Fisher R, Bayir H, Kagan V, Yu J, Liu B, Bahar I, Epperly MW, Wipf P, Wang H, Huq MS, and Greenberger JS. (2020) Anti-Ferroptosis Drug Enhances Total-Body Irradiation Mitigation by Drugs that Block Apoptosis and Necroptosis. Radiat Res 193:435-450. PMID: 32134361 PMCID: PMC7299160
J297. Ponzoni L, Penaherrera DA, Oltvai ZN, and Bahar I (2020) Rhapsody: Predicting the pathogenicity of human missense variants. Bioinformatics 36:3084-3092.PMID: 32101277 PMCID: PMC7214033
J296. Kapralov AA, Yang Q, Dar HH, Tyurina YY, Anthonymuthu TS?, Kim R, St. Croix CM, Mikulska-Ruminska K, Liu B, Shrivastava IH, Tyurin VA, Ting HC, Wu YL, Gao Y, Shurin GV, Artyukhova MA, Ponomareva LA, Timashev8 PS, Domingues RM, Stoyanovsky DA, Greenberger JS, Mallampalli RK, Bahar I, Gabrilovich DI, Bayir H, Kagan VE. (2020) Redox Lipid Reprogramming Commands Susceptibility of Macrophages and Microglia to Ferroptotic Death. Nat Chem Biol 16:278-290. PMID: 32080625 PMCID: PMC7233108
B18. Shrivastava IH, Liu C, Dutta A, Bakan A, Bahar I. (2020) Allostery as Structure-Encoded Collective Dynamics: Significance in Drug Design. Structural Biology in Drug Discovery: Methods, Techniques, and Practices. Eds Jean-Paul Renaud. Chapter 6, p125-141, John Wiley & Sons, NY.
J295. Lee JY, Krieger JM, Li H, Bahar I. (2020) Pharmmaker: Pharmacophore Modeling and Hit Identification Based on Druggability Simulations. Protein Sci 29:76-86 PMID: 31576621 PMCID: 6933858
J294. Zhang S, Chen F, Bahar I. (2020) Differences in the Intrinsic Spatial Dynamics of the Chromatin Contribute to Cell Differentiation. Nucleic Acids Res 48, 1131-1145. PMID: 31828312 PMCID: 7026660
J293. Zhang Y, Doruker P, Kaynak B, Zhang S, Krieger JM, Li H, Bahar I. (2020) Intrinsic dynamics is evolutionarily optimized to enable allosteric behavior. Curr Opin Struct Biol 62:14-21. PMID: 31785465 PMCID: PMC7250723
- Lab Members
Current Lab Members
Anupam Banerjee, PhDAnthony Bogetti, PhDCarlos VenturaDiksha ParwanaHaotian "Frank" ZhangHoang NguyenJiYoung Lee, PhDMary Hongying Cheng, PhDPemra Doruker, PhDRostam Razban, PhDSatyaki SahaSophie LiuXiaowei Bogetti, PhD