J. Peter Gergen, Ph.D.
 SUNY Distinguished Service Professor
SUNY Distinguished Service Professor
Department of Biochemistry and Cell Biology
Center for Developmental Genetics CMM/BLL 112
Stony Brook University
Stony Brook, NY 11794-5140
Office telephone: 631-632-1190
Lab telephone: 631-632-9031
Fax: 631-632-8575
E-mail: john.peter.gergen@stonybrook.edu
- Research Description
 Research in the Gergen laboratory uses the developmental genetic framework of the
Drosophila embryo to investigate the regulation of gene expression, with a primary
focus on the Runt transcription factor. Runt is the founding member of the Runx family
of transcriptional regulators. Runx proteins are present in all animal species examined,
but not in plant or microbial eukaryotic systems. Runt is best characterized for its
role in the Drosophila embryo where it has vital roles in several pathways, including
sex determination, segmentation, and neurogenesis . Mutations in all three human Runx genes are associated with genetic disease, and targeted mutagenesis experiments in
the mouse indicate that these genes have vital roles in several pathways, including
hematopoeisis, neurogenesis and osteogenesis A unifying aspect of Runx function in
these many different pathways is a role in cell fate specification. An intriguing
aspect of regulation by Runt and the vertebrate Runx proteins is that they function
both as transcriptional activators and repressors, depending both on the specific
target gene and the developmental context. Indeed, this context-dependence is central
to the Runt-dependent regulation of the segment-polarity genes during segmentation.
Research in the Gergen laboratory uses the developmental genetic framework of the
Drosophila embryo to investigate the regulation of gene expression, with a primary
focus on the Runt transcription factor. Runt is the founding member of the Runx family
of transcriptional regulators. Runx proteins are present in all animal species examined,
but not in plant or microbial eukaryotic systems. Runt is best characterized for its
role in the Drosophila embryo where it has vital roles in several pathways, including
sex determination, segmentation, and neurogenesis . Mutations in all three human Runx genes are associated with genetic disease, and targeted mutagenesis experiments in
the mouse indicate that these genes have vital roles in several pathways, including
hematopoeisis, neurogenesis and osteogenesis A unifying aspect of Runx function in
these many different pathways is a role in cell fate specification. An intriguing
aspect of regulation by Runt and the vertebrate Runx proteins is that they function
both as transcriptional activators and repressors, depending both on the specific
target gene and the developmental context. Indeed, this context-dependence is central
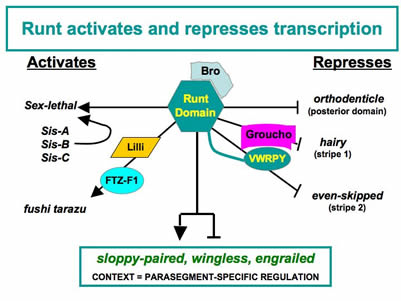
to the Runt-dependent regulation of the segment-polarity genes during segmentation.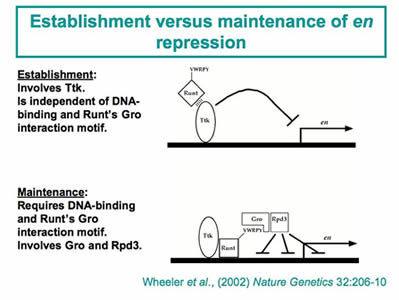 Our recent work has taken advantage of the differential sensitivity of different segment-polarity
gene targets to quantitative manipulations in Runt activity. Runt is a potent repressor
of the segment-polarity geneengrailed. We conducted a genetic screen to identify factors that contribute to this Runt-dependent
repression. Further characterization of the factors identified in this screen revealed
two distinct steps in en repression: 1) an establishment phase that involves a DNA-binding protein encoded
by the tramtrack gene, and 2) a maintenance phase that involves the co-repressor Groucho and the Rpd3
histone de-acetylase. In vivo structure-function studies on Runt confirmed the functional distinction between establishment
and maintenance and further revealed the surprising result that DNA-binding defective
forms of Runt retain the ability to regulate a subset of gene targets. This genetic
screen identified other factors that contribute to Runt’s potency, and ongoing work
aims to further elucidate the roles of these factors in Runt-dependent regulation.
Our recent work has taken advantage of the differential sensitivity of different segment-polarity
gene targets to quantitative manipulations in Runt activity. Runt is a potent repressor
of the segment-polarity geneengrailed. We conducted a genetic screen to identify factors that contribute to this Runt-dependent
repression. Further characterization of the factors identified in this screen revealed
two distinct steps in en repression: 1) an establishment phase that involves a DNA-binding protein encoded
by the tramtrack gene, and 2) a maintenance phase that involves the co-repressor Groucho and the Rpd3
histone de-acetylase. In vivo structure-function studies on Runt confirmed the functional distinction between establishment
and maintenance and further revealed the surprising result that DNA-binding defective
forms of Runt retain the ability to regulate a subset of gene targets. This genetic
screen identified other factors that contribute to Runt’s potency, and ongoing work
aims to further elucidate the roles of these factors in Runt-dependent regulation.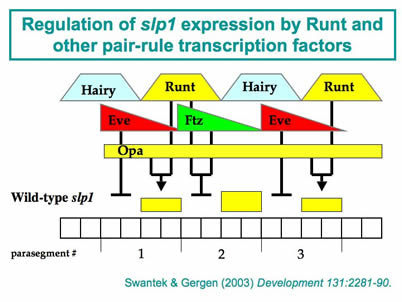 A second major project takes advantage of several attributes of the segment-polarity
target gene sloppy-paired1 (slp1). One key advantage is the simple combinatorial rules that are responsible for modulating
Runt’s activity as both an activator and a repressor of slp1expression. Indeed, using straightforward genetic manipulations it is possible to
achieve uniform, physiologically relevant Runt-dependent activation or repression
of slp1 in all somatic cells of a late blastoderm stage embryo. These embryos provide a platform
for biochemical studies on the in vivoprotein-DNA interactions associated with slp1 repression. The results indicate that the initial establishment of repression does
not involve changes in chromatin re-modeling or modification, nor the assembly of
an initiation complex at the slp1 promoter, but is due instead to developmentally regulated transcriptional elongation.
Ongoing studies are aimed at identifying the specific transcriptional step that is
sensitive to regulation and characterizing the functional contributions of the factor(s)
that are involved. This work is complemented by studies on the slp1 cis-regulatory region that investigate the quantitative contributions of different
cis-regulatory DNA elements to developmentally regulated transcriptional elongation.
A second major project takes advantage of several attributes of the segment-polarity
target gene sloppy-paired1 (slp1). One key advantage is the simple combinatorial rules that are responsible for modulating
Runt’s activity as both an activator and a repressor of slp1expression. Indeed, using straightforward genetic manipulations it is possible to
achieve uniform, physiologically relevant Runt-dependent activation or repression
of slp1 in all somatic cells of a late blastoderm stage embryo. These embryos provide a platform
for biochemical studies on the in vivoprotein-DNA interactions associated with slp1 repression. The results indicate that the initial establishment of repression does
not involve changes in chromatin re-modeling or modification, nor the assembly of
an initiation complex at the slp1 promoter, but is due instead to developmentally regulated transcriptional elongation.
Ongoing studies are aimed at identifying the specific transcriptional step that is
sensitive to regulation and characterizing the functional contributions of the factor(s)
that are involved. This work is complemented by studies on the slp1 cis-regulatory region that investigate the quantitative contributions of different
cis-regulatory DNA elements to developmentally regulated transcriptional elongation. - Publications
Mendoza-García, P., Hugosson, F., Fallah, M., Higgins, M.L., Iwasaki, Y., Pfeifer, K., Wolfstetter, G., Varshney, G., Popichenko, D., Gergen, J.P., Hens, K., Deplancke, B. and Palmer, R.H. (2017) The Zic family homolog Odd-paired regulates Alk expression in Drosophila, PLoS Genetics, 13:e1006617. PMID: 28369060. Abstract
Hang, S. and Gergen, J.P. (2017) Different modes of enhancer-specific regulation by Runt and Eve-skipped during Drosophila segmentation. Mol Biol Cell, Epub Jan 11 , PMID:28077616. Abstract
Walrad PB, Hang S, Gergen JP. (2011) Hairless is a cofactor for Runt-dependent transcriptional regulation. Mol Biol Cell. 22(8):1364-74. doi: 10.1091/mbc.E10-06-0483.Abstract
Wang X, Hang S, Prazak L, Gergen JP. (2010) NELF potentiates gene transcription in the Drosophila embryo. PLoS One 5(7):e11498. doi: 10.1371/journal.pone.0011498.Abstract
Funakoshi Y, Negishi Y, Gergen JP, Seino J, Ishii K, Lennarz WJ, Matsuo I, Ito Y, Taniguchi N, Suzuki T. (2010) Evidence for an essential deglycosylation-independent activity of PNGase in Drosophila melanogaster PLoS One 5(5):e10545. doi: 10.1371/journal.pone.0010545. Abstract
Walrad PB, Hang S, Joseph GS, Salas J, Gergen JP. (2010) Distinct contributions of conserved modules to Runt transcription factor activity. Mol Biol Cell. 21(13):2315-26. doi: 10.1091/mbc.E09-11-0953. Epub 2010 May 12. Abstract
Prazak L, Fujioka M, Gergen JP (2010) Non-additive interactions involving two distinct elements mediate sloppy-paired regulation by pair-rule transcription factors. Dev Biol. 344(2):1048-59. doi: 10.1016/j.ydbio.2010.04.026. Epub 2010 May 6. Abstract
Wang, X., Lee, C., Gilmour, D.S. and Gergen, J.P. (2007) Transcription elongation controls cell specification in the Drosophila embryo, Genes & Dev., 21:1031-6. Abstract
VanderZwan-Butler, C.J., Prazak, L.M. and Gergen, J.P. (2006) The HMG-box protein Lilli is required for Runt-dependent activation of the pair-rule gene fushi tarazu,Developmental Biology, 301:350-360. Abstract
LaLonde, M., Janssens, H., Yun, S., Crosby, J., Redina, O., Olive, V., Altshuller, Y., Du, G., Gergen, J.P. and Frohman, M.A. (2006) A role for Phospholipase D in Drosophila embryonic cellularization, BMC Developmental Biology, 6:60.
Gilbert, M.M., Weaver, B.K., Gergen, J.P. and Reich, N.C. (2005) A novel functional activator of the Drosophila JAK/STAT pathway, Unpaired2, is revealed by an in vivo reporter of pathway activation, Mech Dev. 122:939-48.
LaLonde, M.M., Janssens, H., Rosenbaum, E., Choi, S.-Y., Gergen, J.P., Colley, N.J., Stark, W.S. and Frohman, M.A. (2005) Regulation of phototransduction responsiveness and retinal degeneration by a phospholipase D-generated signaling lipid. J. Cell Biol., 169:471-9.
Van Wijnen, A.J., Stein, G.S., Gergen, J.P., Groner, Y., Hiebert, S.W., Ito, Y., Liu, P., Neil, J.C., Ohki, M. and Speck, N. (2004) Nomenclature for Runt-related (RUNX) proteins. Oncogene 23:4209-10.
Swantek, D. and Gergen, J.P. (2004) Ftz modulates Runt's activity as an activator or repressor of segment-polarity gene transcription, Development 131:2281-90. Abstract
VanderZwan, C., Wheeler, J.C., Li, L.-H., Tracey, W.D., and Gergen, J.P. (2003) A DNA binding-independent pathway of repression by the Drosophila Runt protein. Blood Cells Mol. Dis., 30:207-222. Abstract
Wheeler, John C., Vander Zwan, C., Xu, X., Swantek, D., Tracey, W. D. and Gergen, J.P. (2002) Distinct in vivo requirements for establishment versus maintenance of transcriptional repression, Nature Genetics, 32:206-210. Abstract
Rudolph, M.J. and Gergen, J.P. (2001) DNA-binding by Ig-fold proteins, Nature Structural Biology, 8:384-386
Wheeler, J.C., Shigesada, K, Gergen, J. P. and Ito, Y. (2000) Mechanismsof transcriptional regulation by Runt domain proteins, Seminars in Cell and Developmental Biology, 11:369-375.
Gergen, J.P. (2000) Transcriptional regulation by Runt domain proteins in development and disease, Editorial Perspective (Guest Editor of this review volume), Seminars in Cell and Developmental Biology, 11:325-326.
Tracey, W.D., Ning, X., Klingler, M., Kramer, S.G. and Gergen, J.P. (2000) Quantitative analysis of gene function in the Drosophila embryo, Genetics 154:273-284. Abstract
Li, L.-H. and Gergen, J.P. (1999) Differential interactions between Brother proteins and Runt domain proteins in the Drosophila embryo and eye, Development, 126:3313-3322. Abstract
Wolff, C., Pepling, M., Gergen, P. and Klingler, M. (1999) Structure and evolution of a pair-rule interaction element: runt regulatory sequences in D. melanogaster and D. virilis. Mech. Dev, 80:87-99.
Kramer, S.G., Jinks, T.M., Schedl, P. and Gergen, J.P. (1999) Direct activation of Sex-lethal transcription by the Drosophila Runt protein. Development 126:191-200. Abstract
Tsai, C-C., Kramer, S.G. and Gergen, J.P. (1998) Pair-rule gene runt restricts orthodenticle expression to the presumptive head of the Drosophila embryo. Developmental Genetics 23:35-44. Abstract
Tracey, W.D., Pepling, M.E., Horb, M., Thomsen, G.T. and Gergen, J.P. (1998) A Xenopus homologue of Aml-1 reveals unexpected patterning mechanisms leading to the formation of embryonic blood. Development 125:1371-1380. Abstract
Chen, B., Chu, T., Harms, E., Gergen, J.P. and Strickland, S. (1998) Mapping of Drosophila mutations using site-specific male recombination. Genetics 149:157-163.
Aronson, B.D., Fisher, A.L., Blechman, K., Caudy, M. and Gergen, J.P. (1997) Groucho dependent and independent repression activities of Runt domain proteins. Mol. Cell Biol., 17:5581-5587. Abstract
Klingler, M., Soong, J., Butler, B. and Gergen, J.P. (1996) Disperse versus compact elements for the regulation of runt stripes in Drosophila. Develop. Biol., 177:73-84.Abstract
Lieberfarb, M.E., Chu, T., Wreden, C., Theurkauf, W., Gergen, J.P. and Strickland, S. (1996) Mutations that perturb poly(A)-dependent maternal mRNA activation block the initiation of development. Development, 122:579-588.
Golling, G., Li, L.-H., Pepling, M.E., Stebbins, M. and Gergen, J.P. (1996) Drosophila homologues of proto-oncogene product PEBP2/CBF b regulate the DNA-binding properties of Runt. Mol. Cell Biol., 16:932-942. Abstract
Duffy, J. B., Wells, J. and Gergen, J.P. (1996) Dosage-sensitive maternal modifiers of the segmentation gene runt. Genetics, 142:839-852. Abstract
Pepling, M.E. and Gergen, J.P. (1995) Conservation and function of the transcriptional regulatory protein Runt. Proc. Nat'l. Acad. Sci., USA, 92:9087-9091. Abstract
Tsai, C. and Gergen, J.P. (1995) Pair--rule expression of the Drosophila fushi tarazu gene: a nuclear receptor response element mediates the opposing regulatory effects of runt and hairy. Development, 121:453-462.
Sallés, F.J., Leiberfarb, M.E., Wreden, C., Gergen, J.P. and Strickland, S. (1994) Coordinate initiation of Drosophila development by regulated polyadenylation of maternal mRNAs. Science, 266:1996-1999.
Tsai, C. and Gergen, J.P. (1994) Gap gene properties of the pair-rule gene runt during Drosophila segmentation. Development, 120:1671-1683. Abstract
Duffy, J. B. and Gergen, J.P. (1994) Sex, segments and the CNS: Common mechanisms of cell fate determination. In Advances in Genetics, Vol. 31 (J. Dunlap and J. Hall, eds.), New York, Academic Press, pp. 1-28.
Kagoshima, H., Shigesada, K., Satake, M., Ito, Y., Miyoshi, H., Ohki, M., Pepling, M. and Gergen, J.P. (1993) The Runt-domain identifies a new family of heteromeric transcriptional regulators. Trends in Genetics 9:338-341.
Klingler, M. and Gergen, J.P. (1993) Regulation of runt transcription by Drosophila segmentation genes. Mech. Dev. 43:3-19. Abstract
Butler, B.A., Soong, J. and Gergen, J.P. (1992) The Drosophila segmentation gene runt has an extended cis-regulatory region that is required for vital expression at other stages of development. Mech. Dev. 39:17-28. Abstract
Duffy, J.B. and Gergen, J.P. (1991) The Drosophila segmentation gene runt acts as a position-specific numerator element necessary for the uniform expression of the sex-determining gene Sex-lethal. Genes & Develop. 5:2176 - 2187. Abstract
Duffy, J. B., Kania, M.A., and Gergen, J.P. (1991) Expression and function of the Drosophila gene runt in early stages of neural development. Development 113:1223-1230. Abstract
Kania, M.A., Bonner, A.S., Duffy, J. and Gergen, J.P. (1990) The segmentation gene runt encodes a novel nuclear protein that is also expressed in the developing nervous system. Genes & Develop. 4:1701 - 1713. Abstract
Butler, B.A., Appeddu, P. and Gergen, J.P. (1989). Region-specific mutations of the Drosophila segmentation gene runt. In: Developmental Biology, UCLA Symposia on Molecular and Cellular Biology, New Series, Vol. 125. (E. Davidson, J. Ruderman and J. Posakony, eds.). Alan R. Liss, Inc., New York, New York. pp 105 - 112.
Ingham, P.W. and Gergen, J.P. (1988). Interactions between the pair-rule genes runt, hairy, even-skipped and fushi tarazu generate a molecular prepattern in the Drosophila embryo. (Suppl., Mechanisms of Segmentation) Development 104:51-60.
Gergen, J.P. and Butler, B.A. (1988). Isolation of the Drosophila segmentation gene runt and analysis of its expression during embryogenesis. Genes & Develop. 2:1179-1193. Abstract
Gergen, J.P. (1987). Drosophila segmentation genes and blastoderm cell identities. Bioessays 6:61-66.
Gergen, J.P. (1987). Dosage compensation in Drosophila: Evidence that daughterless and Sex-lethal control X-chromosome activity at the blastoderm stage of embryogenesis. Genetics 117:477-485. Abstract
Tully, T. and Gergen, J.P. (1986). Deletion mapping of the Drosophila memory mutant amnesiac. J. Neurogenetics 3:33-47.
Gergen, J.P., Coulter, D. and Wieschaus, E. (1986). Segmental pattern and blastoderm cell identities. In Gametogenesis and the Early Embryo, (J. Gall, ed.) pp. 195-220. New York: Alan R. Liss.
Gergen, J.P. and Wieschaus, E.F. (1986). Localized requirements for gene activity in segmentation of Drosophila embryos: Analysis of armadillo, fused, giant and unpairedmutations in mosaic embryos. Wilhelm Roux's Arch. 195:49-62.
Gergen, J.P. and Wieschaus, E.F. (1986). Dosage requirements for runt in the segmentation of Drosophila embryos. Cell 45:289-299. Abstract
Zusman, S., Coulter, D. and Gergen, J.P. (1985). Lethal mutations induced in the proximal X-chromosome of Drosophila melanogaster using P-M hybrid dysgenesis. Dros. Inf. Serv. 61:217-218.
Gergen, J.P. and Wieschaus, E.F. (1985). The localized requirements for a gene affecting segmentation in Drosophila: Analysis of larvae mosaic for runt. Develop. Biol. 109:321-335. Abstract
Sharp, W., DeFranco, D., Silberklang, M., Hosbach, H.A., Schmidt, T., Kubli, E., Gergen, J.P., Wensink, P.C. and Soll, D. (1981). The initiator tRNA genes of Drosophila melanogaster: Evidence for a tRNA pseudogene. Nucleic Acids Research, 9:5867-5882.
Gergen, J.P., Loewenberg, J.Y. and Wensink, P.C. (1981). tRNA 2 lys gene clusters in Drosophila. J. Mol. Biol., 147:475-499.
Barnett, T., Pachl, C., Gergen, J.P. and Wensink, P.C. (1980). The isolation and characterization of Drosophila yolk protein genes. Cell 21:729-738.
Gergen, J.P., Stern, R.H. and Wensink, P.C. (1979). Filter replicas and permanent collections of recombinant DNA plasmids. Nucleic Acids Research 7:2115-2136 (1979).Abstract
- Lab Members
- Yasuno IwasakiPostdoc2015-current
- Jennifer ShappMaster's Student2017-current
- Yasuno Iwasaki
- Alumni
Name Position in Gergen lab email location Name Position in Gergen lab email location NameMartin Klingler Position in Gergen labPostdoctoral Associate emailmartin.klingler@fau.de location Dean of Biology, University of Erlangen-NurnbergNameBen Aronson Position in Gergen labPostdoctoral Associate emailBen_Aronson@redlands.edu locationProfessor, University of Redlands NameJohn Wheeler Position in Gergen labPostdoctoral Associate emailjohn_wheeler@sbcglobal.net location Principal Research Scientist, Janssen BioTherapeuticsNameKathy Wojtas Position in Gergen labPostdoctoral Associate emailkathy.wojtas@gmail.com locationVP and Medical Director, BGB Group NameYasuno Iwasaki Position in Gergen labPostdoctoral Associate emailyasuno.iwasaki@stonybrook.edu location Postdoctoral Associate, Stony Brook UniversityName Position in Gergen lab email location NameJoseph Duffy Position in Gergen labDoctoral student emailjduffy@wpi.edu location Head, Department of Biology & Biotechnology, WPINameChih-Cheng Tsai Position in Gergen labDoctoral student emailtsaich@rwjms.rutgers.edu location Associate Professor, Neuroscience and Cell Biology, Rutgers Robert Wood Johnson Medical SchoolNameMelissaa Pepling Position in Gergen labDoctoral student emailmepeplin@syr.edu location Professor and Associate Chair, Department of Biology, Syracuse UniversityNameGreg Golling Position in Gergen labDoctoral student emailggolling@taftcollege.edu locationProfessor and Division Chair, Taft College NameSunita Kramer Position in Gergen labDoctoral student emailsunita.kramer@rutgers.edu location Associate Professor and Associate Dean for Academic Affairs, Honors College, Rutgers UniversityNameLinghui Li Position in Gergen labDoctoral student emaillli@ibms.sinica.edu.tw location Associate Research Scientist, Academica Sinica, TaiwanNameDan Tracey Position in Gergen labDoctoral student emaildtracey@indiana.edu location Associate Professor, Department of Biology, Indiana University BloomingtonNameChristine VanderZwan Position in Gergen labDoctoral student emailchristine.butler@gmail.com locationStay at home mother NamePegine Walrad Position in Gergen labDoctoral student emailpegine.walrad@york.ac.uk location Anniversary Research Lecturer, Department of Biology, University of YorkNameXiaoling Wang Position in Gergen labDoctoral student emailxiaoling.wang@rockefeller.edu location Research Associate, Laboratory of Biochemistry and Molecular Biology, Rockefeller UniversityNameLisa Prazak Position in Gergen labDoctoral student emailprazakl@farmingdale.edu location Assistant Professor, Biology, Farmingdale State CollegeNameSaiyu Hang Position in Gergen labDoctoral student emailsaiyu_hang@hms.harvard.edu location Postdoctoral Associate, Harvard Medical SchoolNameKimberly Bell Position in Gergen labDoctoral student emailkimberly.bell@stonybrook.edu location STEM Postdoctoral Associate, Center for Excellence in Learning & Teaching, Stony BrookNameMichael Higgins Position in Gergen labDoctoral student emailmiluis.higgins@gmail.com location Name Position in Gergen lab email location NameDeborah Swantek Position in Gergen labResearch Assistant emailswantekd@gmail.com location Biology Teacher, Chantilly High School, Fairfax, VAName Position in Gergen lab email location NameRammohan Ramakrishnan Position in Gergen labMasters student emailr_krishna90@yahoo.com location Quality Engineer, Medtronic, Minneapolis-St PaulNameJinelle Wint Position in Gergen labMasters student emailjinelle.wint@stonybrook.edu locationPhD student, Stony Brook University NameJennifer Shapp Position in Gergen labMasters student emailjennifer.shapp@stonybrook.edu locationCurrent student Name Position in Gergen lab email location NameGarfield Clunie, MD Position in Gergen labUndergraduate student emailgarfield.clunie@mssm.edu location Asst. Clinical Professor, Mount Sinai School of MedicineNameHoward Sussman, MD Position in Gergen labUndergraduate student email location Faculty, Family Medicine, Stony Brook University HospitalNameOdeniel Sertil, PhD Position in Gergen labUndergraduate student emailosertil@gmail.com location Forensic Scientist, Phoenix Police Department Crime LaboratoryNameChristopher Magnifico, MD Position in Gergen labUndergraduate student email cmagnifi@sgu.edu locationPhysician, Bayport, NY NameMichael Stebbins, PhD Position in Gergen labUndergraduate student emailMichael.j.stebbins@gmail.com location VP of Science and Technology, Arnold FoundationNameDayna Smith, MD Position in Gergen labUndergraduate student email location Obstetrician-Gynecologist, Fayetteville, GANameSusie Lin, MD Position in Gergen labUndergraduate Honors student emailsusie.lin@vanderbilt.edu location Asst. Professor, Dept. of Oral and Maxillofacial Surgery, Vanderbilty University Medical CenterNameGreg Banninger, PhD Position in Gergen labUndergraduate student emailgbanning@illinois.edu location Patent Coordinator, University of Illinois Urbana-ChampaignNameMary Berketis-Simons Position in Gergen labUndergraduate student email locationChemistry teacher, Seaford High School NameSean Boykevisch, PhD Position in Gergen labUndergraduate student emailsean.boykevisch@stonybrook.edu location Asst. Director (Life Sciences) , Technology Licensing and Industry Relations, Stony Brook ReserchNameGenevieve Jospeh, PhD Position in Gergen labUndergraduate Honors student emailgenevieve.joseph@mssm.edu location Admin. Asst., Department of Oncological Sciences, Icahn School of Medicine at Mount SinaiNameSteven Naymagon, MD Position in Gergen labUndergraduate student emailsteven.naymagon@mountsinai.org location Asst. Professor, Internal Medicine, Icahn School of Medicine at Mount SinaiNameMichael Cressy, PhD Position in Gergen labUndergraduate student emailmcressy@malverne.k12.ny.us location Science Research Teacher, Malverne High SchoolNameJocelin Abraham (Rimes) Position in Gergen labUndergraduate student emailjocelin.rimes@gmail.com locationStay at home mother NameSunita Singh (Celestrin) Position in Gergen labUndergraduate student emailThecelestrins@gmail.com locationStay at home mother NameKevin Celestrin, PhD Position in Gergen labUndergraduate student emailkevin.celestrin@gmail.com location Postdoctoral Associate, Albert Einstein College of MedicineNameGiorgio Medranda, MD Position in Gergen labUndergraduate student emailgionyc86@gmail.com location Cardiovascular Disease Fellow, NYU Winthrop HospitalNameConrad Tenenbaum, PhD Position in Gergen labUndergraduate Honors student emailconrad.tenenbaum@gmail.com location Senior Associate, Blueprint Research Group, Princeton NJNameChristopher Bianco Position in Gergen labUndergraduate student email locationPhD student, NYU School of Medicine NameLiujing Xing, PhD Position in Gergen labUndergraduate Honors student emailxingliujing@gmail.com location Scientific Analyst at Morrison & Forrester LLP, San Francisco, CANameSeema Sawh Position in Gergen labUndergraduate student emailseema.sawh@stemcell.com location Account Manager, STEMCELL Technologies, New York, NYNameAngelita Tan Position in Gergen labUndergraduate student emailangelitatan1@gmail.com location Doctoral Program in Nursing Practice, Columbia UniversityNameKarthink Rao Position in Gergen labUndergraduate student emailmeleegc@yahoo.com location Insurance Agent, Bankers Life, Westborough, MANameTianyu Zhan, PhD Position in Gergen labUndergraduate Honors student emailsewen67@gmail.com location Senior Research Statistician, AbbVie Inc., Chicago, ILNameHaiyue Zhang Position in Gergen labUndergraduate Honors student emailhaiyuezhang222@gmail.com location Biostatistician, Massachusetts General Hospital Biostatistics Center, Boston, MANameMihai Andreca Position in Gergen labUndergraduate student emailmihai.andreca@yahoo.com location Medical student Albert Einstein College of MedicineNameEmily Iocolano Position in Gergen labUndergraduate student emailemily.iocolano6@gmail.com location Veterinary Assistant, The Animal Medical Center, New York, NYNameKevin Skier Position in Gergen labUndergraduate Honors student email location Medical student, University of MassachusettsName Position in Gergen lab email location